The importance of genomics in diagnosis
Haematological malignancies encompass a wide variety of clonal neoplastic disorders arising from haematopoietic and lymphoid tissue. They range from slowly progressing chronic diseases to acute leukaemias and high-grade lymphomas with aggressive clinical courses. The abundance of easily accessible fresh cells and high-quality DNA derived from bone marrow and peripheral blood has always put haematological malignancies at the forefront of the application of genomics.
Haematological malignancies have been well characterised at the genomic level, not only because of the abundance of material, but also as a result of research-driven discoveries.
The importance of genomics in diagnosis, prognostication and management has been recognised for decades, exemplified by the success story of chronic myeloid leukaemia (CML). In the 1960s, Nowell and Hungerford described a small abnormal chromosome in the cells of patients with CML.1 Through improvements in cytogenetic techniques, the ‘Philadelphia chromosome’, resulting from a chromosomal translocation involving the long arms of chromosomes 9 and 22, was identified.
Advances in molecular methodologies led to the discovery that this translocation gave rise to the BCR-ABL1 fusion and a new chimeric protein with enhanced tyrosine kinase function. Almost 40 years later, this abnormal protein was successfully targeted with tyrosine kinase inhibitors, dramatically changing the disease course for patients.2
Haematological malignancies have been well characterised at the genomic level, not only because of the abundance of material, but also as a result of research-driven discoveries. For example, the finding that somatic chromosomal abnormalities – such as translocations t(8;21) in acute myeloid leukaemia (AML), and disease defining t(15;17) in acute promyelocytic leukaemia (APML) – resulted in functional genetic consequences with oncogenic potential.
Much of this knowledge was gained using ‘gold-standard’ cytogenetic techniques, karyotyping and fluorescence in situ hybridisation, and, later, molecular PCR-based assays (Table 1).
| Table 1: Conventional genomic technologies used in clinical management of haematological malignancies. | |
| Method | Description |
| Cytogenetics | The analysis of G-banded chromosomes (karyotyping) to identify structural and numerical abnormalities – translocations, inversions, duplications, aneuploidy, polyploidy. Karyotyping is currently a mandatory component in the diagnostic evaluation of many haematological cancers, including acute leukaemias. Certain chromosome abnormalities are associated with specific diagnoses, prognostic risk stratification and response to treatment. |
| FISH | Use of fluorescently labelled DNA probes to identify the number and location of known DNA sequences within the nucleus. A rapid and more sensitive technique than cytogenetics; however, it is a targeted approach that is best used when specific abnormalities need to be identified/excluded – e.g. rapid confirmation of PML-RARA in cases of suspected APML. |
| PCR | Use of DNA primer sequences to amplify small, targeted regions of the genome for molecular analysis; it is used to detect specific mutations – e.g. FLT3 ITD and TKD mutations in AML, BRAF V600E mutation in hairy cell leukaemia. More sensitive than FISH, rapid and relatively low cost, but is targeted and can only detect specific mutations. |
| Reverse transcription PCR | Uses mRNA copied into cDNA prior to amplification. Used for detecting and quantitating fusion transcripts at high sensitivity, such as monitoring BCR-ABL1 MRD in CML. |
| AML: Acute myeloid leukaemia; APML: Acute promyelocytic leukaemia; CML: Chronic myeloid leukaemia; FISH: Fluorescence in situ hybridisation; MRD: Measurable residual disease. | |
Over the past 20 years, advances in microarray and high-throughput sequencing (HTS) (nextgeneration sequencing) ‘-omics’ technologies have dramatically increased our knowledge of the molecular pathogenesis of haematological malignancies (Table 2).
| Table 2: High-throughput sequencing (next-generation sequencing) methods. | |
| Method | Description |
| Targeted (panel) sequencing | Sets of genes or hotspots of mutation are studied by PCR amplification or oligonucleotide baits that ‘capture’ regions of interest. Provides sensitive and deep coverage of targeted sites, particularly for sequence mutations. The ability to detect rearrangements and copy number alterations is limited by analysis approach. |
| Transcriptome (RNA) sequencing | Transcribed regions of the genome are converted to DNA and sequenced. A powerful and flexible approach that can identify gene rearrangements (RNA fusion panels), sensitive gene-expression profiling, analyse expression of mutations, alternative and novel gene isoform expression, and non-coding RNA expression. |
| Exome sequencing | Regions of the genome corresponding to coding exons (and, variably, additional regions, including promoter regions and non-coding RNAs) are captured and sequenced. DNA copy number alterations and rearrangements that occur in the region of the genes are captured. |
| Whole genome sequencing | The entire DNA of a sample is sequenced. Matched tumour and non-tumour DNA are sequenced and sequence data are mapped to a reference genome. The tumour genome is compared to the matched non-tumour genome to identify somatic variants, and the non-tumour sample is compared with a reference genome to identify germline variants. Coverage is usually greater than exome sequencing (typically 30–50-fold compared with 100–200-fold) and the ability to detect low-frequency, subclonal variants is lower. |
Classifying haematological malignancy and the importance of SIHMDS
Diagnosis and classification of haematological malignancies rely on the integration of morphological, immunophenotypic and genomic information. The Specialised Integrated Haematological Malignancy Diagnostic Service (SIHMDS) is key to this classification, and all SIHMDS now have access to advanced genomic diagnostics. Genetic changes, increasingly in combinations rather than individual lesions, are of vital importance for formal classification as per the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.3
Applications of advanced genomics
Personalised medicine
One of the most important benefits of HTS-based profiling is the identification of specific biomarkers that predict a response, or lack thereof, to both established and novel therapies. Traditional chemotherapeutic options are largely non-targeted, toxic and frequently accompanied by significant morbidity.
Increasingly, focus has shifted to novel (targeted) therapies that are directed at a genetic lesion or associated pathway. A historic example is the use of all-trans retinoic acid in PML-RARA positive APML, which revolutionised outcomes. More recently approved targeted therapies in AML include FLT3 tyrosine kinase inhibitors (quizartinib, midostaurin, gilteritinib) in cases with FLT3 internal tandem duplications and IDH2 inhibitors (enasidenib) in IDH2-mutant AML.
Ultimately, the future clinical management of patients will move away from ‘one size fits all’ chemotherapy and towards a more personalised, genomically targeted approach.
In chronic lymphocytic leukaemia (CLL), patients with TP53 mutations or chromosome 17p deletions have poor prognoses with standard therapy; however, the Bruton’s tyrosine kinase inhibitor ibrutinib has improved outcomes. The ALK inhibitor crizotinib, originally used in nonsmall cell lung cancers, has been approved for paediatric usage and young adults with ALK fusion-positive anaplastic large cell lymphoma.
Ultimately, the future clinical management of patients will move away from ‘one size fits all’ chemotherapy and towards a more personalised, genomically targeted approach. This requires rapid turnaround genetic characterisation and multidisciplinary discussion at genomic tumour boards to assign the significance of mutations and direct appropriate therapies.
Measurable residual disease monitoring
The use of quantitative real-time PCR, and more recently HTS to monitor disease-associated clonal markers post-therapy, has revolutionised how we assess disease response and predict outcomes. The traditional definition of remission is the restoration of normal haematopoiesis with <5% blasts in the bone marrow. This definition has been surpassed by highly sensitive molecular techniques allowing a far deeper assessment of responses. The accurate determination of measurable (formerly minimal) residual disease (MRD) monitoring using immunoglobulin and T-cell receptor genes in acute lymphoblastic leukaemia is the most important independent prognostic biomarker and is used for treatment stratification.4
In AML, MRD is now the standard of care, with monitoring for the most common mutations or gene fusions, such as NPM1, PML-RARA and CBFB-MYH11.5 Monitoring of BCR-ABL1 transcripts by reverse transcription PCR in CML is clearly defined at specific time points for optimal treatment response.2 Longitudinal analysis of blood and bone marrow is being investigated in myeloma and CLL,6,7 while analysis of cell-free DNA (‘liquid biopsies’) is showing promise in lymphoma.8
The future of genomics in haematological malignancies
The NHS has been at the forefront of advances in genomics. Capitalising on the foundations laid by the 100,000 Genomes Project, NHS England has launched a new Genomic Medicine Service. This ambitious new service aims to ensure that all patients in England will have equity of access to advanced, clinically relevant genomic tests while fully embedding the latest genomics technologies in routine practice in the NHS. At the heart of this, for both staff and the public, is genomics education.9
The specific tests available are presented in the National Genomic Test Directory for Cancer and its counterpart for germline disorders, the National Genomic Test Directory for Rare and Inherited Diseases.10 The Directory specifies the genomic tests commissioned, the technology involved and the patients who will be eligible for a test. The national genomic testing service is delivered through a network of seven Genomic Laboratory Hubs, each responsible for coordinating services for a particular part of the country.
The primary aim of WGS is to ensure patients have comprehensive genomic profiling alongside standard of care diagnostics to gain additional information and potentially inform treatments or access to novel therapies and clinical trials.
All patients with acute leukaemia and any paediatric cancer, including all paediatric haematological malignancies, are eligible for whole genome sequencing (WGS). The primary aim of WGS is to ensure patients have comprehensive genomic profiling alongside standard of care diagnostics to gain additional information and potentially inform treatments or access to novel therapies and clinical trials.
WGS coupled with high-quality clinical data will provide a valuable research resource, while also giving patients the opportunity to contribute their genome to the National Genomic Research Library, a collection of data that can be accessed by researchers. As WGS requires the sequencing of both tumour and germline DNA, it also has the potential to identify patients with causal variants in predisposition genes.
Once WGS is of equivalence to existing techniques, and clinically relevant turnaround times become available, it is envisaged that it will replace a number of individual assays with a single test. However, it is acknowledged that the present turnaround times and sensitivity of WGS mean it is unsuitable for replacing very rapid or highsensitivity testing. Other promising technologies include the use of ‘long read’ sequencing, which performs rapid sequencing in real time.11
The future at our fingertips
It is an exciting time to be involved in haematological malignancies, with all these advanced diagnostic technologies at our fingertips. By incorporating the latest genomic advances into routine healthcare, our knowledge and understanding will continue to improve patient outcomes – an example is given below (Figure 1).
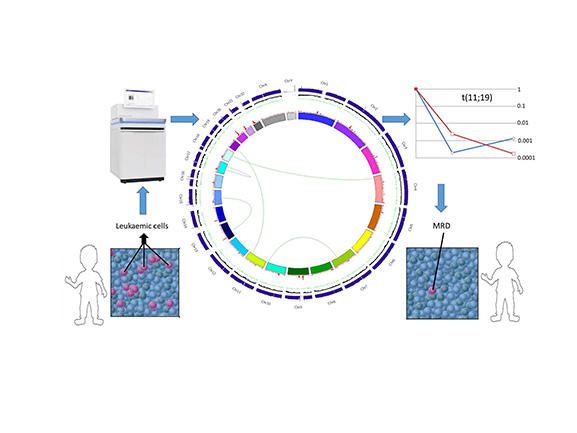
Figure 1: Application of whole genome sequencing in practice.
This schematic shows the application of a whole genome sequence from a patient’s leukaemia. The central plot (a Circos plot) is used to display/visualise the results of whole genome sequencing data on a single patient, which in this case has identified a t(11;19), which is then used as a target for quantitative PCR to track MRD response to therapy over time.
The interpretation of the Circos plot in the centre of the figure is as follows: arcs inside the plot are translocations (green), inversions (purple). Tracks on the Circos plot starting with the innermost track are: 1. Chromosomes 2. Density of somatic SNVs (red) 3. Density of somatic indels (green) 4. Ratio of normalised depth of coverage for tumour vs normal. CNV losses (red), CNV gains (green), copy-neutral LOH regions (yellow) 5. Absolute depth of coverage in tumour sample (blue) (outermost track).
Implementation of these sequencing technologies, along with continuing developments in genomic technology to improve turnaround times, will further reinforce the vital role of genomics in the management not only of haematological patients, but all patients with cancer.