Introduction
Despite significant improvements in cancer management over the last 30 years, mortality rates have remained disappointingly poor for many common cancers, such as those of the colorectum and pancreas.1 The initial expectations that precision oncology would address the poor outcomes for metastatic cancer have been tempered by the challenges associated with tumour heterogeneity, tumour evolution and emergence of resistance mutations.2 This recognition has, in part, renewed interest in exploring opportunities for optimising early detection of cancer and widening targeted therapy, and, in particular, how genomic analysis can contribute to this.
The UK has long been at the forefront of discovery in human genomics and is recognised for its world-leading genetic research, such as UK Biobank and Deciphering Developmental Disorders.
Large-scale sequencing studies such as the International Cancer Genome Consortium and the Cancer Genome Atlas have begun to catalogue the spectra of somatic mutations present in different solid tumours.3 However, to date, there has been minimal traction for solid tumours in aligning large-scale sequencing data to longitudinal data on therapies and outcomes. Stratification using molecular markers could improve the benefit against cost and side effects, which is especially critical in the adjuvant setting. Molecular heterogeneity of tumours and confounding patient factors mean datasets of daunting size will be required. Arguably, these will only be achieved through molecular analyses as standard of care for cancer patients and by adopting a population-level approach to molecular analysis of patients undergoing routine treatments.4
The UK has long been at the forefront of discovery in human genomics and is recognised for its world-leading genetic research, such as UK Biobank and Deciphering Developmental Disorders. Genomics England’s 100,000 Genomes Project was initiated in 2012 to develop infrastructure for routine, high-throughput tumour sequencing (in particular whole-genome sequencing [WGS]) for NHS cancer patients to establish a national research platform of molecular data linked to longitudinal clinical data, and transform delivery of molecular testing in NHS clinical cancer care.5
The 100,000 Genome Project outline
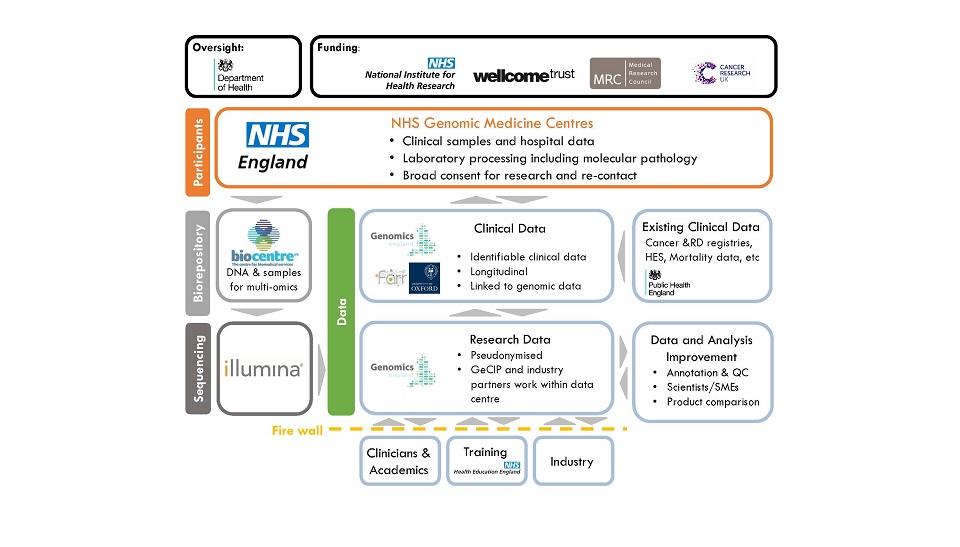
The critical national coverage for the programme was achieved through the creation of 13 NHS Genomic Medicine Centres (GMCs) – hub hospitals linked to up to 18 local recruiting hospitals. Tissue collection and DNA extraction are undertaken locally, followed by transfer of DNA to the central biorepository. WGS of paired DNA (tumour and germline) is then delivered by Illumina at the 100,000 Genomes Sequencing Centre in Hinxton, Cambridgeshire.
A major challenge in the 100,000 Genomes Project was achieving high-quality sequencing while retaining adequate morphology for diagnosis.
Processed sequencing files are then passed back to Genomics England. Using standardised automated pipelines, Genomics England processes the whole-genome analysis. Applying established knowledge, the somatic variants are analysed for potential diagnostic, predictive or prognostic ‘actionability’. Results are presented to the clinical users at GMCs via commercially developed decision-support tools. Supplementary analysis is also supplied, including pan-genomic analyses of tumour signatures and mutational burden.
Clinical legacy: transformation of pathology and establishment of the Genomic Medicine Service
Procurement of tumour DNA of sufficient quantity, quality and purity has been a universal limitation in clinical and research endeavours to date. Formalin fixation and paraffin embedding (FFPE) is the clinical standard in tissue preparation, with fresh tumour tissue being largely the preserve of tissuebanking and research projects. However, formalin causes severe degradation of DNA, affecting the fidelity of the readout in WGS.6 A major challenge in the 100,000 Genomes Project was achieving high-quality sequencing while retaining adequate morphology for diagnosis. Fresh tumour tissue provides much higher quality results, but processing, transporting and storing across diverse settings has been a sizeable challenge.
During the pilot phase of the project, extensive experimental work was undertaken to optimise and standardise DNA extraction from FFPE samples to minimise damage and bring benefit to patients within the NHS.7 Direct comparison of WGS on fresh frozen (FF) versus FFPE tissue found a single nucleotide variant agreement of 71%, while copy number alteration calling was suboptimal in FFPE as a result of non-uniform coverage. The decision was made, therefore, to only accept FF material for WGS.
For many patients, maximum benefit would be gained through analysis of biopsy material, particularly for those for whom surgical resection was not the treatment of choice. Thus, there was a need to establish pathways within the routine clinical setting to acquire biopsies fit for WGS that still fulfil diagnostic requirements and are feasible within a busy service. Many such pathways were established across the GMCs, particularly in colorectal, lung and breast cancers, with 7% of samples sequenced on biopsy pathways. This demonstrates that such pathways are feasible, though this also has resource implications.
Other tumour-specific challenges also exist. For example, with prostate cancer it is sometimes impossible to discern the cancer macroscopically, and, with melanomas, keeping the entire architecture intact is critical for diagnosis. With the likely expansion of WGS in the clinical service, the need for FF pathways, or more economic alternatives, will become more pressing. Exploration of alternative practice, including alternative fixatives, is actively pursued.
Genomic Medicine Service
The 100,000 Genomes Project created the infrastructure for the NHS Genomic Medicine Service, which is delivered by seven genomic laboratory hubs with the aim of providing extended genomic analysis for patients with cancer or rare disease.8 Consistency is facilitated by the National Genomic Test Directory, covering molecular tests for single genes and extended panels through to WGS. The Test Directory will undergo annual review, overseen by the Genomics Clinical Reference Group, but with wide consultation with professional bodies to provide the evidence base for test implementation. Furthermore, all patients will have the opportunity to participate in research contributing to the development of a national genomic knowledge base.
The Airlock is also designed to prevent other providers’ data from being released in a manner that breaches their agreements with Genomics England or their study participants.
Research legacy: Genomics England Research Environment
All analyses of the datasets are carried out within a secure workspace called the Research Environment, to avoid potential security breaches. The Genomics England Research Environment consists of two main platforms, a Linux Virtual Desktop and a HighPerformance Compute (HPC) cluster. The four main types of data are clinical and phenotype, genomic, associated data, and publicly available data. Registered users are provided with access to data with quality metrics (delivered by Illumina) and have access to the Genomics England High Performance Compute Cluster, called Helix, based on IBM’s load sharing facility.
Genomics England does not allow the re-identification of any participant outside the Research Environment – either from the material alone, or through aggregation with other data available now or in the future. The Airlock systems and processes are therefore designed to prevent identifiable data being released from the Research Environment without the consent of the individual concerned.
By adopting a ‘reading library’ approach to data security, researchers can access a richer dataset than otherwise possible. Furthermore, it allows continuous evolution as new datasets become available and the longitudinal datasets expand, without requiring any additional applications from the researchers. The Airlock is also designed to prevent other providers’ data from being released in a manner that breaches their agreements with Genomics England or their study participants.
Clinical data
While a broad dataset was collected for the 100,000 Genomes Project, Genomics England has endeavoured to continue longitudinal clinical data collection, which provides an understanding of the participant’s clinical course. Along with extensive genomic data, the Genomics England dataset is enriched with episodic event and mortality data from NHS Digital, and cancer data derived from the highly curated and focused National Cancer Registration and Analysis Service (NCRAS).
While there is a drive to provide structure and timeliness to clinical data, pathologists and researchers will be keen to explore their own route of experimental interest.
Improving the timeliness and depth of the data available is a particular focus. For instance, reporting of standard of care molecular testing in pathology laboratories is relatively limited within the NCRAS dataset. Genomics England is working in collaboration with NCRAS to explore methods of improving delivery of these results, alongside exploration of more granular data.
Genomics England is keen to explore the utility of minimising the time lag in data curation to improve and diversify the depth of cancer data, and to explore the utility of making de-identified pathology and radiology reports available within the Genomics England Research Environment.
Pathology reports in England have historically been reporter specific, thus each individual pathologist will have their own style and method. There is, however, an increasing drive to have a standardised template for reporting biopsies and excision specimens. Natural language processing (NLP) is extensively used to interpret and analyse large amounts of natural language data. The vast variation in reporting styles can provide unique and interesting challenges in utilising NLP for data mining in pathology reports. Utilising a group of clinical subject matter experts and experienced data scientists, Genomics England has focused on cancer-specific datasets. By augmenting the NCRAScurated data, Genomics England has focused on extracting interesting data points from pathology reports. As a pilot, Genomics England released a colorectal-specific dataset. This included size of tumour, excision margin and microsatellite instability status, in addition to data from NHS Digital and the Office for National Statistics to provide outcome data, including date last seen and date of death.
While there is a drive to provide structure and timeliness to clinical data, pathologists and researchers will be keen to explore their own route of experimental interest. Genomics England has created an academic network, known as the Genomics England Clinical Interpretation Partnership (GeCIP), to develop an understanding of the need to access structured data alongside full reports (Figure 1). To aid this process, Genomics England has developed a process to make a limited number of de-identified pathology and radiology reports available for researchers.
Alongside this wealth of clinical data currently available, pathology and radiology imaging for all cancers will be included alongside primary care data over the next six months to a year. Anyone interested in this data is encouraged to apply to GeCIP, which is organised into ‘domains’ aligning to the different diseases included in the 100,000 Genomes Project.
Molecular diagnostics and the future
Molecular diagnostics and precision medicine are now a reality. With the rollout of the 100,000 Genomes Project, one of the most ambitious initiatives of its kind worldwide, pathways have been established for the adoption of genomic medicine across the NHS. This required a transformation in the way in which the NHS handles patient samples and integrates complementary analyses on tissues to direct patient management. New approaches in molecular pathology are essential to address the challenges of this new era of genomic medicine. Indeed, how pathologists can transit from an approach to tissue diagnostics driven by morphology alone towards a true morphomolecular ethos is arguably the most important test for tissue and cellular pathologists in the last 60 years.9
Beyond the diagnostic setting, there has been renewed interest in tissue pathology in large research environments. There is a clear realisation that effective and robust translational medicine requires large, high-quality, well-annotated tissue samples and expert pathology input. Beyond the importance of accurate histological assessment, pathology can have an impact on translational studies relating to professional biobanking, digital pathology, pathology bioinformatics and information management, and molecular pathological epidemiology. Pathology, and pathologists, are at the centre of genomic medicine, and we need the drive and support to embrace it.